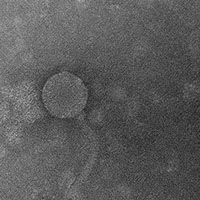
As the final product of the Hampden-Sydney Genomics and Bioinformatics course from the spring 2014 semester, the National Center for Biotechnology Information has accepted the class’ sequence annotation of a bacteriophage discovered at Hampden-Sydney for publication in the GenBank database. The sequence, authored by Josh Dimmick ’15, Grayland Godfrey ’15, William Banning ’15, Mitch Cavallarin ’15, Tommy Isom ’14, Hakeem Mohammed ’14. Jackson Parker ’14, Francis Polakiewicz ’14, Putney Smith ’14, and Professor Mike Wolyniak, was a part of Hampden-Sydney’s participation in the Howard Hughes Medical Institute (HHMI) Science Education Alliance-Phage Hunters Advancing Genomics and Evolutionary Science (SEA-PHAGES) program. Dr. Vassie Ware of Lehigh University collaborated with the final stages of preparing the sequence, which will directly contribute to a national research initiative based out of the University of Pittsburgh that seeks to understand how viruses that infect various bacterial species have evolved over time.
Bacteriophage McFly was isolated by a previous Hampden-Sydney Molecular and Cellular Biology course by Seth Ayers ’11. It is a virus that infects the species Mycobacterium smegmatus and has a genome of approximately 50,000 basepairs. The student authors listed above used several bioinformatics databases over the course of a semester to identify and characterize the predicted genes in the McFly sequence. McFly is the third bacteriophage Hampden-Sydney students have contributed to the national sequence database, joining Arturo and Cheetobro. The SEA-PHAGES initiative allows students from Hampden-Sydney to join undergraduates from across the nation in conducting original research as a component of their scientific training.
To explore McFly, visit the sequence file at https://www.ncbi.nlm.nih.gov/nuccore/1007010572